Publications
For a full list see below or go to Google Scholar
Highlights

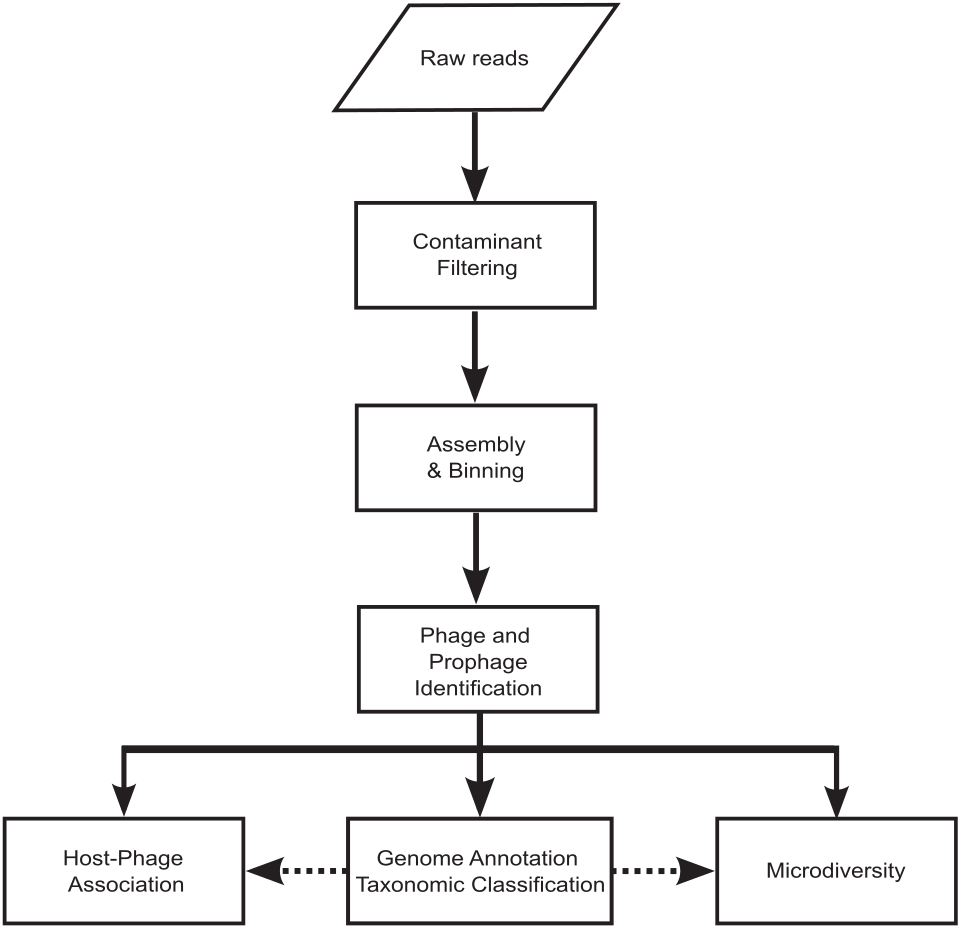
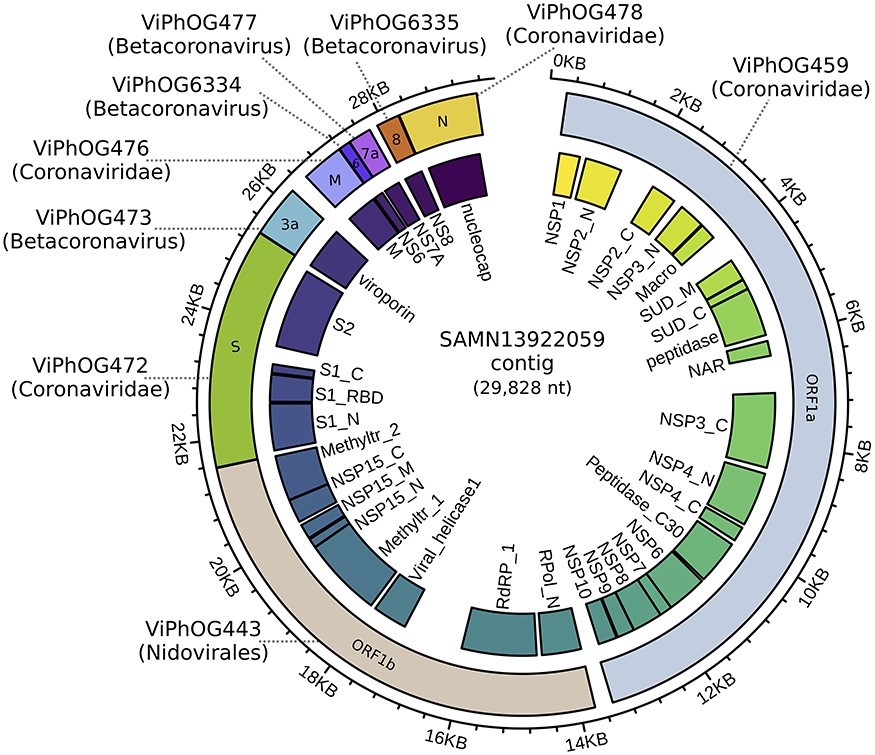
The study introduces VIRify, a computational pipeline for characterizing viral communities based on sequencing data. VIRify accurately identifies viral contigs and prophages, using profile hidden Markov models for taxonomic classification, demonstrating high accuracy in diverse datasets, including microbial mock communities and metagenomic studies.
G Rangel-Pineros, A Almeida, M Beracochea, E Sakharova, M Marz, AR Muñoz, M Hölzer, RD Finn
PLOS Computational Biology 19(8), e1011422 (2023)
This review delves into the significant advancements in bacteriophage research, highlighting the impact of metagenomics in unveiling the vast genetic diversity of phages, leading to the development of computational tools for analyzing phage genome sequences across five key analyses.
JS Andrade-Martínez, LC Camelo Valera, LA Chica Cárdenas, L Forero-Junco, G López-Leal, JL Moreno-Gallego, G Rangel-Pineros, A Reyes
Microbiology and Molecular Biology Reviews 86(2), e00004–21 (2022)
This work explores the emergence of bioinformatics tools tailored for SARS-CoV-2, providing a comprehensive overview of their applications in detecting infections, analyzing sequencing data, tracking the pandemic, studying virus evolution, and developing therapeutic strategies to address the ongoing COVID-19 crisis.
F Hufsky, K Lamkiewicz, A Almeida, A Aouacheria, C Arighi, A Bateman, J Baumbach, N Beerenwinkel, C Brandt, … A Reyes, L Richardson, DL Robertson, S Sadegh, JB Singer, K Theys, C Upton, M Welzel, L Williams, M Marz
Briefings in Bioinformatics 22(2), 642–663 (2021)
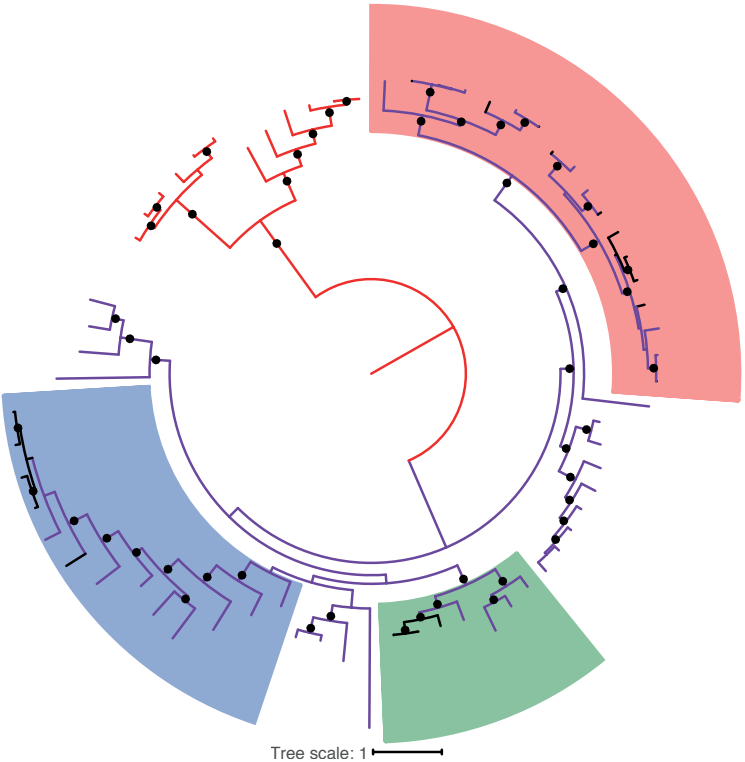
The virome remains a relatively unexplored component of the microbiome. Moreno-Gallego and Chou et al. examined the viromes of monozygotic twins to ask how microbiome diversity relates to virome diversity, without host genetic variables. Twin pairs were sorted by high or low microbiome concordance, which revealed a correlated virome relatedness.
JL Moreno-Gallego, S Chou, SCD Rienzi, JK Goodrich, TD Spector, JT Bell, ND Youngblut, I Hewson, A Reyes, RE Ley
Cell Host & Microbe 25(2), 261–272.e5 (2019)
Full publications list
2023
Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee
D Turner, AN Shkoporov, C Lood, AD Millard, BE Dutilh, P Alfenas-Zerbini, LJ van Zyl, RK Aziz, HM Oksanen, MM Poranen, AM Kropinski, J Barylski, JR Brister, N Chanisvili, RA Edwards, F Enault, A Gillis, P Knezevic, M Krupovic, I Kurtböke, A Kushkina, R Lavigne, S Lehman, M Lobocka, C Moraru, A Moreno Switt, V Morozova, J Nakavuma, A Reyes Muñoz, J Rūmnieks, B Sarkar, MB Sullivan, J Uchiyama, J Wittmann, T Yigang, EM Adriaenssens
Archives of Virology 168(2), 74 (2023)
Four principles to establish a universal virus taxonomy
P Simmonds, EM Adriaenssens, FM Zerbini, NGA Abrescia, P Aiewsakun, P Alfenas-Zerbini, Y Bao, J Barylski, C Drosten, S Duffy, WP Duprex, BE Dutilh, SF Elena, ML García, S Junglen, A Katzourakis, EV Koonin, M Krupovic, JH Kuhn, AJ Lambert, EJ Lefkowitz, M Łobocka, C Lood, J Mahony, JP Meier-Kolthoff, AR Mushegian, HM Oksanen, MM Poranen, A Reyes-Muñoz, DL Robertson, S Roux, L Rubino, S Sabanadzovic, S Siddell, T Skern, DB Smith, MB Sullivan, N Suzuki, D Turner, KV Doorslaer, A Vandamme, A Varsani, N Vasilakis
PLOS Biology 21(2), e3001922 (2023)
Rational Design of Profile HMMs for Sensitive and Specific Sequence Detection with Case Studies Applied to Viruses, Bacteriophages, and Casposons
LS Oliveira, A Reyes, BE Dutilh, A Gruber
Viruses 15(2), 519 (2023)
Bacteriophages playing nice: Lysogenic bacteriophage replication stable in the human gut microbiota
SG Sutcliffe, A Reyes, CF Maurice
iScience 26(2) (2023)
Genomic and Evolutionary Features of Nine AHPND Positive Vibrio parahaemolyticus Strains Isolated from South American Shrimp Farms
A Castellanos, L Restrepo, L Bajaña, I Betancourt, B Bayot, A Reyes
Microbiology Spectrum 11(4), e04851–22 (2023)
Comparative Genomics Identifies Conserved and Variable TAL Effectors in African Strains of the Cotton Pathogen Xanthomonas citri pv. malvacearum
AL Pérez-Quintero, LM Rodriguez-R, S Cuesta-Morrondo, E Hakalová, D Betancurt-Anzola, LCC Valera, LAC Cardenas, L Matiz-Céron, JM Jacobs, V Roman-Reyna, AR Muñoz, AJB Giraldo, R Koebnik
Phytopathology® 113(8), 1387–1393 (2023)
VIRify: An integrated detection, annotation and taxonomic classification pipeline using virus-specific protein profile hidden Markov models
G Rangel-Pineros, A Almeida, M Beracochea, E Sakharova, M Marz, AR Muñoz, M Hölzer, RD Finn
PLOS Computational Biology 19(8), e1011422 (2023)
Guidelines for public database submission of uncultivated virus genome sequences for taxonomic classification
EM Adriaenssens, S Roux, JR Brister, I Karsch-Mizrachi, JH Kuhn, A Varsani, T Yigang, A Reyes, C Lood, EJ Lefkowitz, MB Sullivan, RA Edwards, P Simmonds, L Rubino, S Sabanadzovic, M Krupovic, BE Dutilh
Nature Biotechnology 41(7), 898–902 (2023)
Conserved, yet disruption-prone, gut microbiomes in neotropical bumblebees
N Villabona, N Moran, T Hammer, A Reyes
mSphere 8(6), e00139–23 (2023)
2022
Comparing novel shotgun DNA sequencing and state-of-the-art proteomics approaches for authentication of fish species in mixed samples
MS Varunjikar, C Moreno-Ibarguen, JS Andrade-Martinez, H Tung, I Belghit, M Palmblad, PA Olsvik, A Reyes, JD Rasinger, KK Lie
Food Control 131, 108417 (2022)
The gut microbiota of chickens in a commercial farm treated with a Salmonella phage cocktail
V Clavijo, T Morales, MJ Vives-Flores, A Reyes Muñoz
Scientific Reports 12(1), 991 (2022)
Taxonomical Evaluation of Plant Chloroplastic Markers by Bayesian Classifier
L Matiz-Ceron, A Reyes, J Anzola
Frontiers in Plant Science 12 (2022)
Functional and Phylogenetic Characterization of Bacteria in Bovine Rumen Using Fractionation of Ruminal Fluid
R Hernández, M Chaib De Mares, H Jimenez, A Reyes, A Caro-Quintero
Frontiers in Microbiology 13 (2022)
Analysis and exploration of microbial traits in a wet coffee fermentation experiment using MGnify
A Peña González, A Reyes Muñoz
F1000Research 11(401), 401 (2022)
Computational Tools for the Analysis of Uncultivated Phage Genomes
JS Andrade-Martínez, LC Camelo Valera, LA Chica Cárdenas, L Forero-Junco, G López-Leal, JL Moreno-Gallego, G Rangel-Pineros, A Reyes
Microbiology and Molecular Biology Reviews 86(2), e00004–21 (2022)
Mining of Thousands of Prokaryotic Genomes Reveals High Abundance of Prophages with a Strictly Narrow Host Range
G López-Leal, LC Camelo-Valera, JM Hurtado-Ramírez, J Verleyen, S Castillo-Ramírez, A Reyes-Muñoz
mSystems 7(4), e00326–22 (2022)
2021
Bacterial meta-analysis of chicken cecal microbiota
LAC Cardenas, V Clavijo, M Vives, A Reyes
PeerJ 9, e10571 (2021)
Computational strategies to combat COVID-19: useful tools to accelerate SARS-CoV-2 and coronavirus research
F Hufsky, K Lamkiewicz, A Almeida, A Aouacheria, C Arighi, A Bateman, J Baumbach, N Beerenwinkel, C Brandt, M Cacciabue, S Chuguransky, O Drechsel, RD Finn, A Fritz, S Fuchs, G Hattab, A Hauschild, D Heider, M Hoffmann, M Hölzer, S Hoops, L Kaderali, I Kalvari, M von Kleist, R Kmiecinski, D Kühnert, G Lasso, P Libin, M List, HF Löchel, MJ Martin, R Martin, J Matschinske, AC McHardy, P Mendes, J Mistry, V Navratil, EP Nawrocki, ÁN O’Toole, N Ontiveros-Palacios, AI Petrov, G Rangel-Pineros, N Redaschi, S Reimering, K Reinert, A Reyes, L Richardson, DL Robertson, S Sadegh, JB Singer, K Theys, C Upton, M Welzel, L Williams, M Marz
Briefings in Bioinformatics 22(2), 642–663 (2021)
Genome Sequence and Characterization of Lactobacillus casei Phage, vB_LcaM_Lbab1 Isolated from Raw Milk
V Gradaschi, F Payaslian, ME Dieterle, L Rondón Salazar, E Urdániz, M Di Paola, J Peña Cárcamo, F Zon, M Allievi, E Sosa, D Fernandez Do Porto, M Dunne, P Goeller, J Klumpp, RR Raya, A Reyes, M Piuri
PHAGE 2(1), 57–63 (2021)
Tightening Bonds in Latin America Through Phage Discovery
F Payaslian, V Gradaschi, L Rondón Salazar, ME Dieterle, E Urdániz, M Di Paola, F Zon, M Allievi, C Sanchez Rivas, RR Raya, A Reyes, M Piuri
PHAGE 2(1), 7–10 (2021)
Isolation and Characterization of vB_MsmS_Celfi: A New Mycobacterium tuberculosis Bacteriophage
F Payaslian, V Gradaschi, L Rondón Salazar, ME Dieterle, E Urdániz, M Di Paola, J Peña Cárcamo, F Zon, E Sosa, D Fernandez Do Porto, M Dunne, P Goeller, J Klumpp, RR Raya, A Reyes, M Piuri
PHAGE 2(1), 43–49 (2021)
Monitoring the variation in the gut microbiota of captive woolly monkeys related to changes in diet during a reintroduction process
C Quiroga-González, LAC Cardenas, M Ramírez, A Reyes, C González, PR Stevenson
Scientific Reports 11(1), 6522 (2021)
A novel vieuvirus from multidrug-resistant Acinetobacter baumannii
G López-Leal, A Reyes-Muñoz, RI Santamaria, MA Cevallos, C Pérez-Monter, S Castillo-Ramírez
Archives of Virology 166(5), 1401–1408 (2021)
Discovery of new potential CDK2/VEGFR2 type II inhibitors by fragmentation and virtual screening of natural products
AF Vásquez, A Reyes Muñoz, J Duitama, A González Barrios
Journal of Biomolecular Structure and Dynamics 39(9), 3285–3299 (2021)
Informative Regions In Viral Genomes
JL Moreno-Gallego, A Reyes
Viruses 13(6), 1164 (2021)
Disentangling the Complexity of the Rumen Microbial Diversity Through Fractionation Using a Sucrose Density Gradient
R Hernández, H Jimenez, C Vargas-Garcia, A Caro-Quintero, A Reyes
Frontiers in Microbiology 12 (2021)
Challenges and Considerations for Delivering Bioinformatics Training in LMICs: Perspectives From Pan-African and Latin American Bioinformatics Networks
V Ras, P Carvajal-López, P Gopalasingam, A Matimba, PA Chauke, N Mulder, F Guerfali, VD Del Angel, A Reyes, G Oliveira, J De Las Rivas, M Cristancho
Frontiers in Education 6 (2021)
Non-Extensive Fragmentation of Natural Products and Pharmacophore-Based Virtual Screening as a Practical Approach to Identify Novel Promising Chemical Scaffolds
AF Vásquez, A Reyes Muñoz, J Duitama, A González Barrios
Frontiers in Chemistry 9 (2021)
Practical metagenomics: The Study of human gut microbiome in health and disease: Applications in Acute Diarrheal Diseases (ADD)
A Peña González, A Reyes Muñoz, A Madrigal Leiva
F1000Research 10(861), 861 (2021)
Bacterial Viruses Subcommittee and Archaeal Viruses Subcommittee of the ICTV: update of taxonomy changes in 2021
M Krupovic, D Turner, V Morozova, M Dyall-Smith, HM Oksanen, R Edwards, BE Dutilh, SM Lehman, A Reyes, DP Baquero, MB Sullivan, J Uchiyama, J Nakavuma, J Barylski, MJ Young, S Du, P Alfenas-Zerbini, A Kushkina, AM Kropinski, I Kurtböke, JR Brister, C Lood, BL Sarkar, T Yigang, Y Liu, L Huang, J Wittmann, N Chanishvili, LJ van Zyl, J Rumnieks, T Mochizuki, M Jalasvuori, RK Aziz, M Łobocka, KM Stedman, AN Shkoporov, A Gillis, X Peng, F Enault, P Knezevic, R Lavigne, S Rhee, V Cvirkaite-Krupovic, C Moraru, AI Moreno Switt, MM Poranen, A Millard, D Prangishvili, EM Adriaenssens
Archives of Virology 166(11), 3239–3244 (2021)
A whole genome duplication drives the genome evolution of Phytophthora betacei, a closely related species to Phytophthora infestans
DA Ayala-Usma, M Cárdenas, R Guyot, MCD Mares, A Bernal, AR Muñoz, S Restrepo
BMC Genomics 22(1), 795 (2021)
Microbial community characterization of shrimp survivors to AHPND challenge test treated with an effective shrimp probiotic (Vibrio diabolicus)
L Restrepo, C Domínguez-Borbor, L Bajaña, I Betancourt, J Rodríguez, B Bayot, A Reyes
Microbiome 9(1), 88 (2021)
Perspective on taxonomic classification of uncultivated viruses
BE Dutilh, A Varsani, Y Tong, P Simmonds, S Sabanadzovic, L Rubino, S Roux, AR Muñoz, C Lood, EJ Lefkowitz, JH Kuhn, M Krupovic, RA Edwards, JR Brister, EM Adriaenssens, MB Sullivan
Current Opinion in Virology 51, 207–215 (2021)
From Trees to Clouds: PhageClouds for Fast Comparison of ∼640,000 Phage Genomic Sequences and Host-Centric Visualization Using Genomic Network Graphs
G Rangel-Pineros, A Millard, S Michniewski, D Scanlan, K Sirén, A Reyes, B Petersen, MR Clokie, T Sicheritz-Pontén
PHAGE 2(4), 194–203 (2021)
Ten simple rules for organizing a bioinformatics training course in low- and middle-income countries
B Moore, P Carvajal-López, PA Chauke, M Cristancho, VDD Angel, SL Fernandez-Valverde, A Ghouila, P Gopalasingam, FZ Guerfali, A Matimba, SL Morgan, G Oliveira, V Ras, A Reyes, JDL Rivas, N Mulder
PLOS Computational Biology 17(8), e1009218 (2021)
2020
International consensus definition of low anterior resection syndrome
C Keane, NS Fearnhead, L Bordeianou, P Christensen, E Espin Basany, S Laurberg, A Mellgren, C Messick, GR Orangio, A Verjee, K Wing, I Bissett, tLIC Group
Colorectal Disease 22(3), 331–341 (2020)
Fine Resolution Analysis of Microbial Communities Provides Insights Into the Variability of Cocoa Bean Fermentation
ME Pacheco-Montealegre, LL Dávila-Mora, LM Botero-Rute, A Reyes, A Caro-Quintero
Frontiers in Microbiology 11 (2020)
Genome-Scale Metabolic Model of Xanthomonas phaseoli pv. manihotis: An Approach to Elucidate Pathogenicity at the Metabolic Level
D Botero, J Monk, MJ Rodríguez Cubillos, A Rodríguez Cubillos, M Restrepo, V Bernal-Galeano, A Reyes, A González Barrios, BØ Palsson, S Restrepo, A Bernal
Frontiers in Genetics 11 (2020)
Paving the Way to Unveil the Diversity and Evolution of Phage Genomes
A Reyes, MJ Vives
Viruses 12(9), 905 (2020)
Diet Quality, Food Groups and Nutrients Associated with the Gut Microbiota in a Nonwestern Population
ÁS García-Vega, V Corrales-Agudelo, A Reyes, JS Escobar
Nutrients 12(10), 2938 (2020)
Essentials in metagenomics (Part I)
A Peña González, A Reyes Muñoz
F1000Research 9(1194), 1194 (2020)
Essentials in Metagenomics (Part II)
A Peña González, A Reyes Muñoz
F1000Research 9(1195), 1195 (2020)
2019
Minimum Information about an Uncultivated Virus Genome (MIUViG)
S Roux, EM Adriaenssens, BE Dutilh, EV Koonin, AM Kropinski, M Krupovic, JH Kuhn, R Lavigne, JR Brister, A Varsani, C Amid, RK Aziz, SR Bordenstein, P Bork, M Breitbart, GR Cochrane, RA Daly, C Desnues, MB Duhaime, JB Emerson, F Enault, JA Fuhrman, P Hingamp, P Hugenholtz, BL Hurwitz, NN Ivanova, JM Labonté, K Lee, RR Malmstrom, M Martinez-Garcia, IK Mizrachi, H Ogata, D Páez-Espino, M Petit, C Putonti, T Rattei, A Reyes, F Rodriguez-Valera, K Rosario, L Schriml, F Schulz, GF Steward, MB Sullivan, S Sunagawa, CA Suttle, B Temperton, SG Tringe, RV Thurber, NS Webster, KL Whiteson, SW Wilhelm, KE Wommack, T Woyke, KC Wrighton, P Yilmaz, T Yoshida, MJ Young, N Yutin, LZ Allen, NC Kyrpides, EA Eloe-Fadrosh
Nature Biotechnology 37(1), 29–37 (2019)
Toward unrestricted use of public genomic data
RI Amann, S Baichoo, BJ Blencowe, P Bork, M Borodovsky, C Brooksbank, PSG Chain, RR Colwell, DG Daffonchio, A Danchin, V de Lorenzo, PC Dorrestein, RD Finn, CM Fraser, JA Gilbert, SJ Hallam, P Hugenholtz, JPA Ioannidis, JK Jansson, JF Kim, H Klenk, MG Klotz, R Knight, KT Konstantinidis, NC Kyrpides, CE Mason, AC McHardy, F Meyer, CA Ouzounis, AAN Patrinos, M Podar, KS Pollard, J Ravel, AR Muñoz, RJ Roberts, R Rosselló-Móra, S Sansone, PD Schloss, LM Schriml, JC Setubal, R Sorek, RL Stevens, JM Tiedje, A Turjanski, GW Tyson, DW Ussery, GM Weinstock, O White, WB Whitman, I Xenarios
Science 363(6425), 350–352 (2019)
Virome Diversity Correlates with Intestinal Microbiome Diversity in Adult Monozygotic Twins
JL Moreno-Gallego, S Chou, SCD Rienzi, JK Goodrich, TD Spector, JT Bell, ND Youngblut, I Hewson, A Reyes, RE Ley
Cell Host \& Microbe 25(2), 261–272.e5 (2019)
Defining a Core Genome for the Herpesvirales and Exploring their Evolutionary Relationship with the Caudovirales
JS Andrade-Martínez, JL Moreno-Gallego, A Reyes
Scientific Reports 9(1), 11342 (2019)
Bogotá River anthropogenic contamination alters microbial communities and promotes spread of antibiotic resistance genes
CE Posada-Perlaza, A Ramírez-Rojas, P Porras, B Adu-Oppong, A Botero-Coy, F Hernández, JM Anzola, L Díaz, G Dantas, A Reyes, MM Zambrano
Scientific Reports 9(1), 11764 (2019)
Global phylogeography and ancient evolution of the widespread human gut virus crAssphage
RA Edwards, AA Vega, HM Norman, M Ohaeri, K Levi, EA Dinsdale, O Cinek, RK Aziz, K McNair, JJ Barr, K Bibby, SJJ Brouns, A Cazares, PA de Jonge, C Desnues, SL Díaz Muñoz, PC Fineran, A Kurilshikov, R Lavigne, K Mazankova, DT McCarthy, FL Nobrega, A Reyes Muñoz, G Tapia, N Trefault, AV Tyakht, P Vinuesa, J Wagemans, A Zhernakova, FM Aarestrup, G Ahmadov, A Alassaf, J Anton, A Asangba, EK Billings, VA Cantu, JM Carlton, D Cazares, G Cho, T Condeff, P Cortés, M Cranfield, DA Cuevas, R De la Iglesia, P Decewicz, MP Doane, NJ Dominy, L Dziewit, BM Elwasila, AM Eren, C Franz, J Fu, C Garcia-Aljaro, E Ghedin, KM Gulino, JM Haggerty, SR Head, RS Hendriksen, C Hill, H Hyöty, EN Ilina, MT Irwin, TC Jeffries, J Jofre, RE Junge, ST Kelley, M Khan Mirzaei, M Kowalewski, D Kumaresan, SR Leigh, D Lipson, ES Lisitsyna, M Llagostera, JM Maritz, LC Marr, A McCann, S Molshanski-Mor, S Monteiro, B Moreira-Grez, M Morris, L Mugisha, M Muniesa, H Neve, N Nguyen, OD Nigro, AS Nilsson, T O’Connell, R Odeh, A Oliver, M Piuri, AJ Prussin II, U Qimron, Z Quan, P Rainetova, A Ramírez-Rojas, R Raya, K Reasor, GAO Rice, A Rossi, R Santos, J Shimashita, EN Stachler, LC Stene, R Strain, R Stumpf, PJ Torres, A Twaddle, M Ugochi Ibekwe, N Villagra, S Wandro, B White, A Whiteley, KL Whiteson, C Wijmenga, MM Zambrano, H Zschach, BE Dutilh
Nature Microbiology 4(10), 1727–1736 (2019)
Computational search for UV radiation resistance strategies in Deinococcus swuensis isolated from Paramo ecosystems
J Díaz-Riaño, L Posada, IC Acosta, C Ruíz-Pérez, C García-Castillo, A Reyes, MM Zambrano
PLOS ONE 14(12), e0221540 (2019)
2018
Holobiont Diversity in a Reef-Building Coral over Its Entire Depth Range in the Mesophotic Zone
FL Gonzalez-Zapata, P Bongaerts, C Ramírez-Portilla, B Adu-Oppong, G Walljasper, A Reyes, JA Sanchez
Frontiers in Marine Science 5 (2018)
Draft Genome Sequence of a White Spot Syndrome Virus Isolate Obtained in Ecuador
L Restrepo, A Reyes, L Bajaña, I Betancourt, B Bayot
Genome Announcements 6(26), 10.1128/genomea.00605–18 (2018)
PirVP genes causing AHPND identified in a new Vibrio species (Vibrio punensis) within the commensal Orientalis clade
L Restrepo, B Bayot, S Arciniegas, L Bajaña, I Betancourt, F Panchana, A Reyes Muñoz
Scientific Reports 8(1), 13080 (2018)
Local confinement of disease-related microbiome facilitates recovery of gorgonian sea fans from necrotic-patch disease
E Quintanilla, C Ramírez-Portilla, B Adu-Oppong, G Walljasper, SP Glaeser, T Wilke, AR Muñoz, JA Sánchez
Scientific Reports 8(1), 14636 (2018)
2017
Advances in Gut Microbiome Research, Opening New Strategies to Cope with a Western Lifestyle
GP Rodriguez-Castaño, A Caro-Quintero, A Reyes, F Lizcano
Frontiers in Genetics 7 (2017)
Captivity Shapes the Gut Microbiota of Andean Bears: Insights into Health Surveillance
A Borbón-García, A Reyes, M Vives-Flórez, S Caballero
Frontiers in Microbiology 8 (2017)
Use of profile hidden Markov models in viral discovery: current insights
A Reyes, J Marcelo P Alves, AM Durham, A Gruber
Advances in Genomics and Genetics 7, 29–45 (2017)
Changing a Generation’s Way of Thinking: Teaching Computational Thinking Through Programming
F Buitrago Flórez, R Casallas, M Hernández, A Reyes, S Restrepo, G Danies
Review of Educational Research 87(4), 834–860 (2017)
Editorial: Virus Discovery by Metagenomics: The (Im)possibilities
BE Dutilh, A Reyes, RJ Hall, KL Whiteson
Frontiers in Microbiology 8 (2017)
2016
GenSeed-HMM: A Tool for Progressive Assembly Using Profile HMMs as Seeds and its Application in Alpavirinae Viral Discovery from Metagenomic Data
JMP Alves, AL de Oliveira, TOM Sandberg, JL Moreno-Gallego, MAF de Toledo, EMM de Moura, LS Oliveira, AM Durham, DU Mehnert, PMdA Zanotto, A Reyes, A Gruber
Frontiers in Microbiology 7 (2016)
2015
Gut DNA viromes of Malawian twins discordant for severe acute malnutrition
A Reyes, LV Blanton, S Cao, G Zhao, M Manary, I Trehan, MI Smith, D Wang, HW Virgin, F Rohwer, JI Gordon
Proceedings of the National Academy of Sciences 112(38), 11941–11946 (2015)
2014
Bacteria from Diverse Habitats Colonize and Compete in the Mouse Gut
H Seedorf, NW Griffin, VK Ridaura, A Reyes, J Cheng, FE Rey, MI Smith, GM Simon, RH Scheffrahn, D Woebken, AM Spormann, W Van Treuren, LK Ursell, M Pirrung, A Robbins-Pianka, BL Cantarel, V Lombard, B Henrissat, R Knight, JI Gordon
Cell 159(2), 253–266 (2014)
HCoDES Reveals Chromosomal DNA End Structures with Single-Nucleotide Resolution
Y Dorsett, Y Zhou, AT Tubbs, B Chen, C Purman, B Lee, R George, AL Bredemeyer, J Zhao, E Sodergen, GM Weinstock, ND Han, A Reyes, EM Oltz, D Dorsett, Z Misulovin, JE Payton, BP Sleckman
Molecular Cell 56(6), 808–818 (2014)
2013
Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing
ES Lim, A Reyes, M Antonio, D Saha, UN Ikumapayi, M Adeyemi, OC Stine, R Skelton, DC Brennan, RS Mkakosya, MJ Manary, JI Gordon, D Wang
Virology 436(2), 295–303 (2013)
Gnotobiotic mouse model of phage–bacterial host dynamics in the human gut
A Reyes, M Wu, NP McNulty, FL Rohwer, JI Gordon
Proceedings of the National Academy of Sciences 110(50), 20236–20241 (2013)
2012
A Noncomplementation Screen for Quantitative Trait Alleles in Saccharomyces cerevisiae
HS Kim, J Huh, L Riles, A Reyes, JC Fay
G3 Genes{\textbar}Genomes{\textbar}Genetics 2(7), 753–760 (2012)
The Shared Antibiotic Resistome of Soil Bacteria and Human Pathogens
KJ Forsberg, A Reyes, B Wang, EM Selleck, MOA Sommer, G Dantas
Science 337(6098), 1107–1111 (2012)
Going viral: next-generation sequencing applied to phage populations in the human gut
A Reyes, NP Semenkovich, K Whiteson, F Rohwer, JI Gordon
Nature Reviews Microbiology 10(9), 607–617 (2012)
Identification of MW Polyomavirus, a Novel Polyomavirus in Human Stool
EA Siebrasse, A Reyes, ES Lim, G Zhao, RS Mkakosya, MJ Manary, JI Gordon, D Wang
Journal of Virology 86(19), 10321–10326 (2012)
IS-seq: a novel high throughput survey of in vivo IS6110 transposition in multiple Mycobacterium tuberculosis genomes
A Reyes, A Sandoval, A Cubillos-Ruiz, KE Varley, I Hernández-Neuta, S Samper, C Martín, MJ García, V Ritacco, L López, J Robledo, MM Zambrano, RD Mitra, P Del Portillo
BMC Genomics 13(1), 249 (2012)
2011
Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice
AL Goodman, G Kallstrom, JJ Faith, A Reyes, A Moore, G Dantas, JI Gordon
Proceedings of the National Academy of Sciences 108(15), 6252–6257 (2011)
2010
Viruses in the faecal microbiota of monozygotic twins and their mothers
A Reyes, M Haynes, N Hanson, FE Angly, AC Heath, F Rohwer, JI Gordon
Nature 466(7304), 334–338 (2010)
2009
Differential gene expression in White Spot Syndrome Virus (WSSV)-infected naïve and previously challenged Pacific white shrimp Penaeus (Litopenaeus) vannamei
JC García, A Reyes, M Salazar, CB Granja
Aquaculture 289(3), 253–258 (2009)
2007
Temperature modifies gene expression in subcuticular epithelial cells of white spot syndrome virus-infected Litopenaeus vannamei
A Reyes, M Salazar, C Granja
Developmental \& Comparative Immunology 31(1), 23–29 (2007)
Mutations in DNA repair genes are associated with the Haarlem lineage of Mycobacterium tuberculosis independently of their antibiotic resistance
J Olano, B López, A Reyes, M del Pilar Lemos, N Correa, P Del Portillo, L Barrera, J Robledo, V Ritacco, M Mercedes Zambrano
Tuberculosis 87(6), 502–508 (2007)